Recent & Upcoming Seminars
2025
Designing Reinforcement Learning Algorithms for Digital Interventions: Pre-Implementation Guidelines
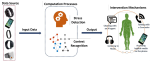
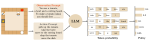
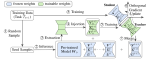
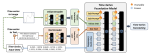
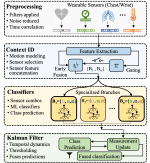
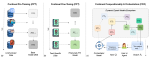
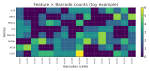
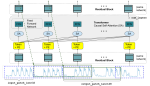
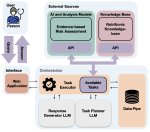
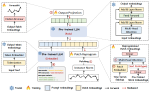
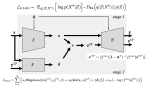
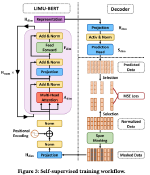
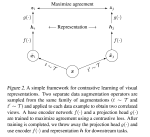
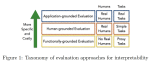
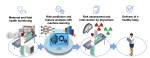
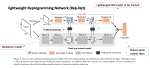
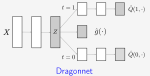
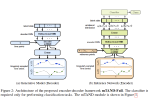
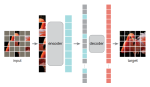
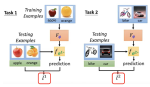
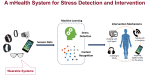
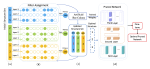
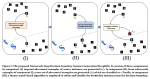
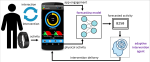
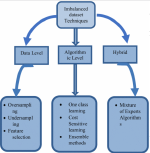
This presentation examines the extension of the PCS (Predictability, Computability, Stability) framework for Reinforcement Learning (RL) algorithm design in digital interventions. Since RL models must be finalized before deployment, robust pre-implementation planning is critical. A case study on Oralytics, a digital oral health intervention, demonstrates how PCS principles enhance RL design. Using a contextual bandit algorithm with Bayesian Linear Regression (BLR), the system optimized intervention effectiveness while ensuring stability. Results showed that BLR performed best, larger clusters improved learning, and simulations required greater diversity. Overall, PCS offers a systematic approach for RL-driven interventions, balancing personalization, computational feasibility, and stability. Further refinements are needed to enhance adaptability and generalization.